Single molecule microscopy to analyze protein dynamics and distributions

Our laboratory uses cutting edge microscopy techniques to study T cell activation in space and time.
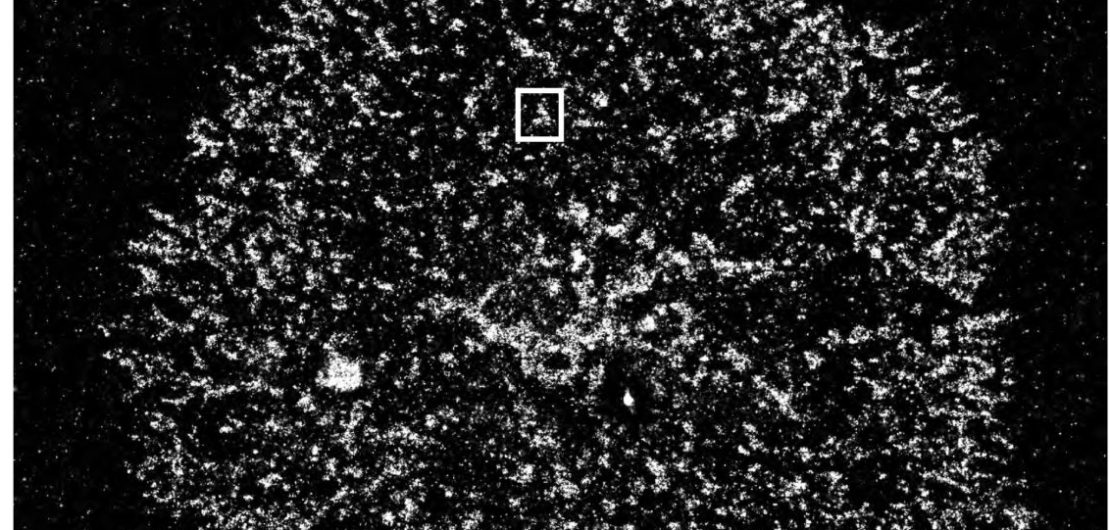
Multicolor or multiplexing super-resolution microscopy allows to visualize the distribution of T cell signaling molecules within the plasma membrane. We have found that T cell signaling molecules are pre-organized and segregated into nanometer sized plasma membrane domains, called nanoclusters or protein islands. Upon T cell activation nanoclusters concatenate to form larger microclusters without fusing or mixing of their content. Microclusters move to the center of the contact site between T cells and antigen presenting cells to form the immunological synapse. We are currently investigating how signaling is controlled by this unique membrane organization and how regulatory signals, e.g. from the inhibitory PD-1 pathway, are spatially and dynamically integrated.
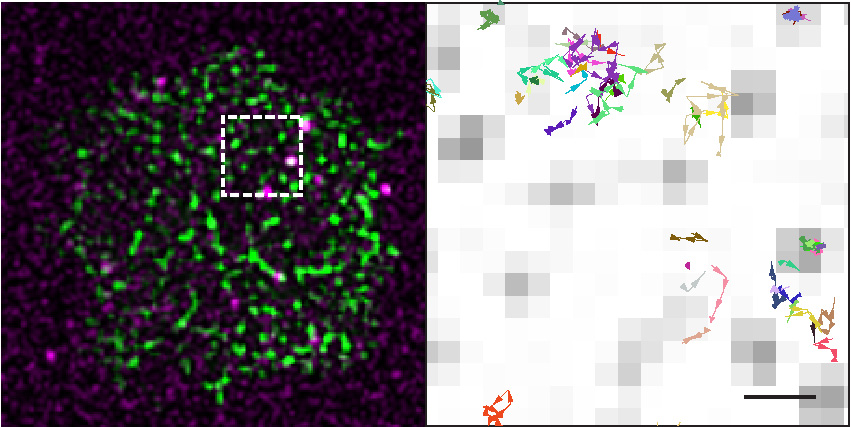
Single molecule tracking is used to study the movement and receptor interactions of signaling molecules at the T cell plasma membrane, specifically of the tyrosine kinase Zap70 and the phosphatase SHP2. Combining this approach with mutant analyses enables us to elucidate molecular mechanisms that control the activation of the enzymes and the translocation of signals within the plane of the plasma membrane. For example, we have found that Zap70 is activated through a cycle of T cell receptor (TCR) binding, activation and release that amplifies antigenic stimuli and allows signal transfer between membrane nanodomains. We continue to study the membrane dynamics of Zap70 and determine if similar mechanisms are utilized by other T cell signaling molecules.
In addition, we are developing novel multiplexing labeling techniques based on Points Accumulation for Imaging in Nanoscale Topography (PAINT) to allow higher spatial resolution and quantitative microscopy. These methods improve the analyses of relative distributions of signaling molecules within the plasma membrane. Our next goal for these methods is to relate single molecule dynamics to the overall organization of the plasma membrane at nanometer resolution.